Mission and Goals
Data science and algorithms for cancer research
The Institute for Computational Cancer Biology (ICCB) at the University Hospital Cologne was founded in 2022 to advance cancer research through computational methods and to train and educate the next generation of computational cancer scientists.
We are part of the Cancer Research Center Cologne Essen (CCCE) funded by the Ministry of Culture and Science of the State of North Rhine-Westphalia.
Our mission is to develop bespoke statistical methods, machine learning approaches, algorithms and models to decipher tumour heterogeneity and cancer evolution and improve our understanding of the wealth of genomic, transcriptomic, epigenomic and imaging data collected in cancer research.
Want to know more?
| ▶ Research spotlights | ▶ Work with us |
About us
News
April 2, 2024
News & Views in Nature Genetics
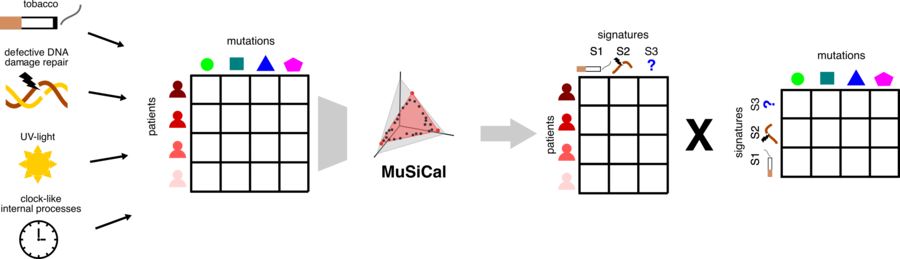
Roland and Tom were invited to give their views on the Nature Genetics paper of Jin et al. presenting MuSiCal, a set of novel algorithms for improved identification of cancer mutational processes by mutational signatures.
October 25, 2022
Multiple job openings in the Schwarzlab
The Schwarzlab at the ICCB has multiple job openings for PhD students, Postdoctoral Fellows and a Scientific Programmer in multiple national and international projects working on chromosomal instability, copy-number evolution and algorithms for tumour heterogeneity.
September 20, 2022
The Lehmann Lab joins the ICCB
We are glad that the Lehmann Lab has joined the ICCB as an associated group.
The Lehmann Lab is interested in the development of approaches that support the molecular characterisation of patient cohorts and in approaches for data integration to gain insights into disease mechanisms. Welcome on board!
Find out more
Research Spotlights

Schwarz Lab - Cologne/Berlin
Cancer Genomics and Evolution
Algorithms for inferring and simulating cancer evolution and for understanding tumour heterogeneity, with a special focus on chromosomal instability and somatic copy-number alterations.

Lehmann Lab - Aachen
Molecular Signatures and Data Integration
Approaches that support the molecular characterisation of patient cohorts and reveal disease mechanisms through integration of diverse types of molecular data.
Sponsors and Partners


Funder
University Hospital and University of Cologne



Funder
Cancer Research Center Cologne Essen (CCCE)
funded by the Ministry of Culture and Science of the State of North Rhine-Westphalia.



Strategic partner
Berlin Institute for the Foundations of Learning and Data (BIFOLD)
funded by the German Ministry for Research and Education (BMBF).



