April 2, 2024
News & Views in Nature Genetics
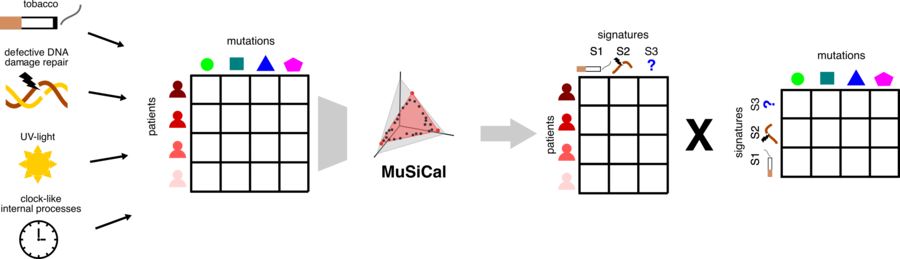
Roland and Tom were invited to give their views on the Nature Genetics paper of Jin et al. presenting MuSiCal, a set of novel algorithms for improved identification of cancer mutational processes by mutational signatures.
November 9, 2023
MOP-C paper published in Blood

As part of a multidisciplinary team led by the Borchmann Lab the Schwarzlab implemented a publicly available shiny application for MOP-C (Molecular prognostic index for central nervous system lymphomas).
MOP-C provides risk assessment for central nervous system lymphomas based on clinical risk factors, radiographic response and peripheral residual disease measured by circulating tumor DNA (Heger et al. 2023, Blood).
By integrating these clinical and molecular features MOP-C was proven to be highly predictive of outcomes a CNSL cohort with a failure-free survival hazard ratio (HR) per risk group of 6.60.
October 23, 2023
Refphase paper published in PLOS Computational Biology
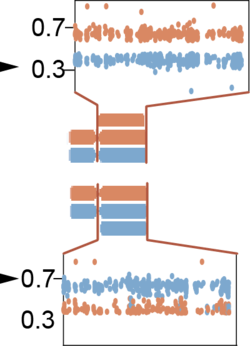
We are excited that after 9 years of development, our multi-region copy-number phasing algorithm Refphase is finally here!
Refphase uses WES or WGS data from multiple samples from the same patients to identify the haplotypes of origin of somatic copy-number alterations (SCNAs). Refphase has led to the discovery of the Mirrored Subclonal Allelic Imbalance (MSAI) phenomenon in cancer (Jamal-Hanjani et al. 2017, Nature), where SCNAs affect opposite haplotypes in the same tumour, leading to a "mirrored" B-allele frequency pattern.
Refphase was further key to demonstrating widespread MSAI occurrences and continuous parallel evolution across human cancers (Watkins et al. 2020, Nature).
Refphase is implemented in R and available on Bitbucket at https://bitbucket.org/schwarzlab/refphase.
May 18, 2023
Marina Petkovic successfully defends her PhD

We are thrilled to report the third successful PhD defense in the Schwarzlab: Marina Petkovic defended her PhD on May 12, 2023 at the Berlin Institute for Molecular Systems Biology, Max Delbrück Center for Molecular Medicine in the Helmholtz Association.
Her PhD thesis is titled "Reconstructing the evolutionary history of cancer from allele-specific copynumber profiles" and she will receive her PhD from the Humboldt University Berlin.
During her PhD she contributed in a major way to the development of MEDICC2 and conducted large scale analyses on the order of evolutionary copy-number events in cancer.
After an interim postdoc at Charite Berlin, Marina is now moving to greener pastures in industry where she will continue to work in bioinformatics.
May 7, 2023
Our team is growing!



The Schwarzlab is welcoming two new PhD students at our site in Cologne at the ICCB and one postdoc at BIFOLD Berlin!
Claudia Robens is a biotechnologist who did her undergrad and Masters in Heidelberg. She will be investigating chromatin architecture in cancer in a collaboration with Ana Pombo from Berlin and as part of the DFG priority program SPP2202.
Katyayni Ganesan is a biologist turned bioinformatician with a Bachelors from the University of Nottingham and a Masters from LMU Munich. She will be investigating single-cell heterogeneity and fitness effects in cancer as part of the SATURN3 consortium.
Nathan Lee is a new postdoctoral fellow at BIFOLD Berlin and a mathematician from the University of Washington. He will be investigating theoretical models of cancer evolution and algorithms for phasing structural variants in single cells.
February 24, 2023
SMITH paper published in Bioinformatics
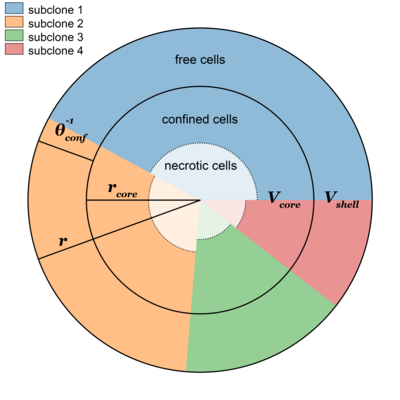
We are thrilled to anounce the release of the SMITH software toolkit for simulating cancer evolution with spatial constraints and the accompanying paper in Bioinformatics.
SMITH is a novel tool and algorithm for simulating cancer evolution including random mutations with varying fitness effects. SMITH is based on the classic branching model of cancer which it extends with local and global "confinement", new mechanisms which simulates spatial constraints by limiting clonal growth based on the size of clones and the overall tumour.
SMITH is implemented in C# and available on Bitbucket at https://bitbucket.org/schwarzlab/smith.
It's accompanying visualisation package pyFish is implemented in Python and available at https://bitbucket.org/schwarzlab/pyfish.
January 31, 2023
Stella de Biase successfully defends her PhD

With flying colors Stella de Biase defended her PhD on January 10, 2023 at the Berlin Institute for Molecular Systems Biology, Max Delbrück Center for Molecular Medicine in the Helmholtz Association.
Her PhD thesis is titled "Exploring the contribution of genetic and environmental factors to cancer risk and development" and she will receive her PhD from the Humboldt University Berlin.
After her Masters degree in Medical Biotechnologies at the Federico II University of Naples, Italy, Stella joined our lab in January 2017 on a MDC NYU PhD Fellowship. Some highlights of her work include the early detection of lung cancer, the effect of SSR mutations on growth rates in yeast, and how smoking affects the expression of SARS-CoV-2 entry genes.
Go Stella! Thanks for all your hard work and kind words. You rock!
November 14, 2022
MEDICC2 manuscript published in Genome Biology!
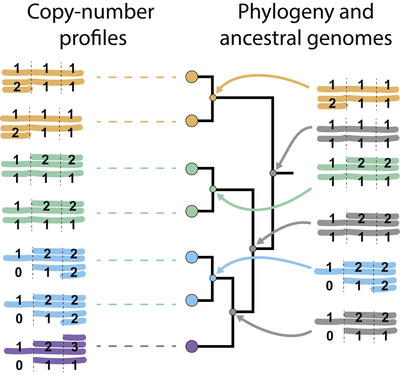
Eight years after its intial release we are more than proud to announce that the MEDICC2 algorithm and paper for inferring cancer evolution from somatic copy-number alterations is now published online at Genome Biology.
MEDICC2 is the leading algorithm for phylogenetic reconstruction from and quantification of chromosomal instability. It is based on an efficient finite-state transducer framework which, together with integrated parallelisations, allows accurate phylogentic inference even for single-cell data with thousands of cells.
MEDICC2 not only outperforms the original implementation in speed by orders of magnitude, but also, as the first and only algorithm in the field, allows the accurate detection of clonal and subclonal whole-genome doubling events.
November 9, 2022
Welcome to our new lab members!




The Schwarzlab is welcoming several new members to our group at our site in Cologne at the ICCB!
Daniel Schütte is a clinician scientist who is interested in molecular biomarkers and cancer early detection and prevention and already joined some months ago.
Felix Schifferdecker and Selina Wächter are two MD students who will be writing their dissertation in the lab in parallel to their MD coursework and clinical pratice over the next years. Felix will be working on simulating chromosomal aberrations in cancer evolution and Selina will be investigating methods for detecting selection in cancer phylogenies.
Finally, we would like to welcome Teodora Bucaciuc, who has joined the lab as a research associate, developing pipelines for copy-number analysis and investigating the co-evolution of genomic and epignomic alterations in cancer. Welcome all, great to have you on board!
October 21, 2022
Julia Markowski successfully defends her PhD

We are incredibly thrilled that Julia Markowski successfully defended her PhD on October 21, 2022 at the Berlin Institute for Molecular Systems Biology, Max Delbrück Center for Molecular Medicine in the Helmholtz Association.
Her PhD thesis is titled "Inferring haplotype-specific chromatin conformation using Genome Architecture Mapping" and she will receive her PhD from the Humboldt University Berlin in the near future.
After completing her Masters degree in Bioinformatics at the Max-Planck-Institute for Molecular Genetics and the University of Potsdam, Julia joined our lab in May 2017 on a MDC PhD Fellowship. She was subsequently supported by the DFG priority programme SPP2202 "Chromatin Architecture in Health and Disease".
She is now moving to Boston as a postdoctoral fellow in the Park Lab at Harvard Medical School and we wish her all the best for the future!



